Family history- a major risk factor for prostate cancer development
The identification of germline mutations in hereditary cancers is of great importance. These mutations make it possible to identify individuals who are at high risk for developing the disease (e.g. BRCA1/2 in breast cancer) and to develop targeted therapeutic approaches directed at events that drive tumor initiation and progression (e.g. renal cancers).
For this reason, for the last three decades this has been an area of intense investigation at the Brady Institute. Today, based on twin studies, we know that prostate cancer is more heritable than other common cancers, including ovarian, kidney, breast and colon cancer.
Paradoxically, although the genetics of these less heritable malignancies is well established, there is still much that is unknown about the genetic pathogenesis of prostate cancer. For this reason, I selected this topic and thought I would tell the story of our search for the missing heritability of the disease.
Family history is a major risk factor
The story begins in 1987 when a 49-year-old man asked me if prostate cancer was hereditary. When I asked him why he wanted to know he responded that his father, his father’s three brothers, and his grandfather died from the disease. At that time, it was common knowledge that women with a mother or sister who had breast cancer had a two-fold higher risk for developing it. So why didn’t I know the answer to his question. Because the available literature on prostate cancer was sparse. There was information from the Utah Mormon genealogical registry but it was unclear if these findings could be applied to the general population or whether they represented a rare founder mutation in an isolated population of men. To determine if the same were true in another population we undertook a case-control study of 742 consecutive men who underwent a radical prostatectomy at Hopkins using their spouses or female companions as a control. Our findings, which confirmed a 2.2 fold increased risk for men with one first-degree relative, were soon confirmed by four other studies that reported similar results.
Familial aggregation is genetic
The next step was to determine whether this familial aggregation was caused by inherited genetic factors or from shared environment. Using the same population, we performed a segregation analysis that demonstrated the best model predicted autosomal dominant inheritance of a rare (0.3%) high penetrance risk allele in families with early age of diagnosis and multiple affected family members. This study demonstrated for the first time that prostate cancer is inherited in Mendelian fashion and provided a foundation for gene mapping studies of heritable prostate cancer. Finally, based on the segregation analysis we developed a definition for hereditary prostate cancer (HPC): three or more first-degree relatives (father, son, or brother); or three generations (grandfather, father, son); or two first-degree relatives if both were less than 55 years old at the time of diagnosis.
Two major genetic influences: Monogenetic and complex
It has been estimated that 5%-10% of prostate cancer may be considered hereditary. Armed with this information we set out with the hope of finding these offending mutations, a journey that has taken over 20 years and is not yet finished. I will pause for a minute to describe the two major categories of genetic influences. Monogenic disease is caused by mutations that are rare but have high penetrance in causing the disease, like the breast cancer genes BRCA1/2. In contrast, complex disease is caused by common genetic variants (single nucleotide polymorphisms, SNPs) that have low penetrance, and for this reason it takes many of them to cause disease, like Type 2 diabetes mellitus.
Linkage analysis to identify loci for rare high penetrance alleles
Initially, we used linkage analysis to search for the rare high penetrant genes. Linkage analysis is used to identify the chromosomal location of a gene by finding DNA sequences of known chromosomal location that are consistently co-inherited with the disease in multiplex families. Within two years we’re pleased to identify the first prostate cancer susceptibility gene on chromosome 1 (1q24-25) but disappointed when our findings could not be replicated by other investigators. Soon, linkage was reported by others at numerous other loci but again few were consistently confirmed and none fulfilled the criteria for a high penetrance allele like BRCA1/2.
Genome-wide association studies to search for complex disease
Next, investigators became excited about the influence of SNPs, and the possibility that a large number with each having a very small effect would be the answer. These studies led to the successful identification of over 100 SNPs associated with prostate cancer risk.
Although the relative increase in risk for any single SNP is small, the risk increases as the number of inherited risk SNPs increases but these appear to explain only about 25% of the risk associated with a positive family history and the clinical utility of these findings remains uncertain.
Where is the missing heritability – rare variants?
So where is the missing heritability? Going back to the drawing board, but this time armed with high- throughput next-generation sequencing (NGS), we again searched for rare variants with the hope that there may be multiple rare variants that contribute a large effect.
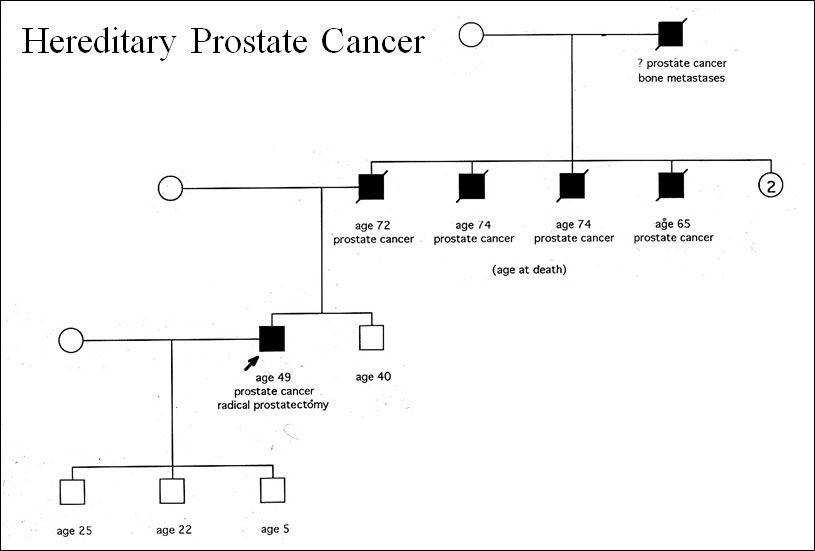
Figure 1: Schematic diagram in a case-control study of men with a history of hereditary prostate cancer and radical prostatectomy
HOXB13
Our first use of NGS was in collaboration with investigators at the University of Michigan who had reported linkage at 17q21. We identified one particular missense variant mutation in a homeobox gene called HOXB13, changing glycine to glutamine at codon 84 or G84E. We were especially interested in this gene as it was already known to be prostate-specific in its expression pattern, and in mice it plays a critical role in the development and maintenance of normal prostate function. This time confirmatory studies involving many thousands of men unequivocally established HOX B13 as the first validated prostate cancer susceptibility gene. Clinically, this mutation is particularly important to be aware of in men with Swedish or Finnish ancestry.
BRCA1/2, DNA Repair, and Mismatch repair mutations Twenty years ago BRCA2 was implicated as an important gene for prostate cancer by studies in Icelandic families and more recently Eeles and her research group at the Royal Marsden provided additional evidence that defective BRCA2 genes are associated with inherited risk of more aggressive prostate cancer. However, it was only within the last couple of years that we learned of their impact on the development of castration-resistant prostate cancer (CRPC).
A study by the Step up to Cancer research group, which carried out the first in-depth sequencing of men with CRPC, demonstrated that 6% of men with CRPC had deleterious germline mutations of BRCA2. When coupled with other genes involved in DNA repair (like ATM, and the mismatch repair genes in Lynch syndrome, e.g. MSH2) the total number of CRPC patients with inherited mutations rose to over 12%. In a study at Hopkins, together with our colleagues at NorthSHore, we found that in men who died from prostate cancer prior to age 65, 10 to 12% carried mutations in BRCA2/BRCA1 and ATM.
Have we been looking in the wrong place?
No one knows for sure, but based on the recent discovery that patients with lethal and aggressive prostate cancer who do not have a strong family history can carry DNA repair mutations in their germline, maybe we have been looking at the wrong patients. Our studies have always concentrated on men with multiple affected family members who are alive. Instead, if future studies concentrate on patients with lethal and advanced disease it is possible we will uncover many previously unknown important pathways.
Clinical implications
Family history is a major risk factor for development of the disease. The history should include age at diagnosis of prostate cancer in both paternal and maternal lineages and a complete list of other cancers. Factors suggestive of a genetic contribution to prostate cancer include the following: 1) multiple affected
first-degree relatives with prostate cancer, including three successive generations with prostate cancer in the maternal or paternal lineage; 2) early onset prostate cancer (age 55 years); or 3) prostate cancer with a family history of the BRCA1/2 mutation or other cancers (e.g., breast, ovarian, pancreatic).
Who should be referred to a genetic counselor for genetic testing? First: men of Nordic descent are up five to 10 times more likely to carry a high risk HOXB13 allele, and thus men in this group should be offered screening for mutations in this gene. Men testing positive should be counseled about earlier and more intensive disease screening.
Second, men with a family history of the BRCA1/2 mutation. Also there is consideration to extend testing to men where the likelihood of a BRCA 1/2 mutation is high: family history of a first-degree relative with breast/ovarian/pancreatic cancer or personal history of breast cancer. Third: men with CPRC. They may benefit from genetic testing utilizing a panel of DNA repair genes for the impact it can have on their individual treatment options (the use of platinum and PARP inhibitors rather than taxanes), as well as information to be shared with family members regarding risk of prostate cancer and other BRCA2 related cancers, e.g. pancreas, breast, ovarian.
In closing, it is important to emphasize that this effort has been led by Dr. William Isaacs who has dedicated his skill, intellect, and energy for the last three decades to uncover the genetic pathogenesis of prostate cancer. This talk and these discoveries are a tribute to his brilliance and perseverance.
Monday 27 March
10.30-10.50: Plenary session 05, Update on hereditary prostate cancer
Author:
Dr. Patrick C. Walsh University Distinguished Service Professor Emeritus The James Buchanan Brady Urological Institute
Baltimore, Maryland (USA)

