Limitations of PSMA: Can DDR biomarkers be used to guide PSMA-directed therapies?
 By Dr. Pasquale Rescigno (London, GB)
By Dr. Pasquale Rescigno (London, GB)
This article reflects the highlights of the lecture Dr. Rescigno gave at the EAU20 Virtual Congress on Friday 17 July. His presentation can be found in the EAU20 Resource Centre.
Prostate-specific membrane antigen (PSMA) is a type II membrane protein. It is expressed in healthy prostate and kidney tissues, salivary glands, nervous system glia, and the small bowel jejunal brush border, but also in prostatic adenocarcinoma (PC)1-4. The PSMA gene is located on the short arm of chromosome 11, a region uncommonly deleted in PC5.
The PSMA protein consists of 3 portions (external, transmembrane, and internal)6. It acts as a glutamate-preferring carboxy-peptidase7 and plays a role in the cellular uptake of folate and glutamate, which are key for the synthesis and repair of DNA. Interestingly, its expression seems to be inversely related to androgen levels8.
The first anti-PSMA antibody that was developed (mAb 7E11) can recognise and bind a PSMA intracellular and/or cytoplasmic epitope1. From a clinical perspective, more than 90% of metastatic castration-resistant PC (mCRPC) patients presented PSMA-positive lesions based on 68Ga-PSMA positron emission tomography/computed tomography (PET/CT) images9. The detection rate of relapse and occult metastatic disease is similarly high in patients with biochemical reoccurrence post-radical treatment and PSA > 2ng/L10. However, ~ 50% of mCRPC patients do not respond to PSMA radioligand therapy despite documented PSMA expression11,12.
Membranous PSMA protein expression
In our recent work, we evaluated membranous PSMA (mPSMA) protein expression in mCRPC biopsies and, when available, the matching, same-patient, diagnostic, castration-sensitive prostate cancer (CSPC) biopsies by immunohistochemistry (IHC), using a mouse monoclonal Ab internally validated. Membranous PSMA quantification for each sample was determined by a pathologist blinded to clinical and molecular data using modified H-scores. The overall percentage of mPSMA positivity across the entire stained tumour sample was determined, yielding a range from 0 to 30013.
Prognostic role of PSMA expression
We first studied mPSMA expression levels in 38 CSPC PC diagnostic biopsies (median H-score 17.5 [0.0–60.0]). While 16 (42%) patient samples had no detectable expression, the remaining 22 expressed PSMA, with a significant degree of interpatient and intrapatient heterogeneity in expression. In our analyses, higher levels of mPSMA expression were associated with a higher Gleason grade (p = 0.04) and a worse overall survival (hazard ratio [95% CI]: H-score < 17.5 = 1.00 vs H-score > 17.5 = 2.97 [1.38–6.43]; p = 0.006).
Expression of mPSMA protein demonstrates intra- and interpatient heterogeneity
Median mPSMA H-score was 55.0 (2.8–117.5) in 60 mCRPC samples. Importantly, 16 (27%) of these biopsies had no detectable expression of mPSMA. Interestingly, a comparison with matched, same patient CSPC tissue samples revealed that half of these samples with no mPSMA protein expression (8/16) were also negative for mPSMA expression at PC diagnosis.
Overall, 37 of the 44 mCRPC mPSMA+ samples (84%) demonstrated marked heterogeneity in expression. This also depended on type of tissue biopsy analysed, with liver metastases (n = 8) showing overall lower PSMA expression compared to bone (n = 25), lymphoid tissue (n = 22), or other sites (n = 5).
mPSMA expression was heterogeneous within the same patient’s sample, with regions of PSMA-negative tumour cells segregated from regions of PSMA expressing cells, a number of which were > 2 mm apart, which is the maximal tissue penetrance of the beta particle emitter 177Lu used in current clinical trials. Furthermore, heterogeneous mPSMA expression between metastases from the same patient was also observed (see figure 1A).
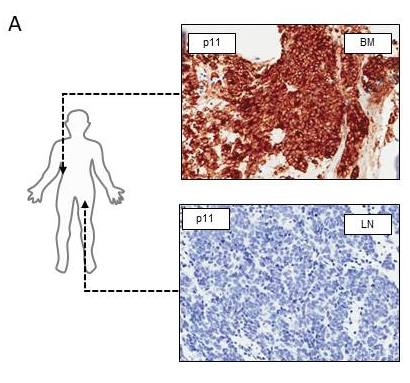
Figure 1A: Bone and lymph node mCRPC biopsies from the same patient demonstrating significant heterogeneity between metastases
Expression of mPSMA protein in mCRPC and DNA repair
In addition to this, considering that intratumour heterogeneity can be significantly increased by genomic instability14, we have also pursued next-generation sequencing (NGS) studies to investigate whether deleterious DNA damage repair (DDR) aberrations were associated with PSMA protein expression.
Overall, mCRPC samples from patients harbouring DDR aberrations had significantly higher (p = 0.016) mPSMA expression than those without these aberrations (DDR 87.5 [25.0–247.5] vs no DDR 20 [0.3–98.8]; difference in medians 60 [5.0–95.0].
More interestingly, when we compared the change in mPSMA protein expression between matched CSPC and mCRPC tissue samples, we observed that mPSMA expression increased significantly in mCRPC patients with detectable DDR aberrations (p = 0.02), but not in those without detectable DDR gene mutations (p = 0.19) (Coef = 35; 95% CI: 2–68; p = 0.04; see figure 1C).
When we explored the association between PSMA expression and DNA repair using RNA-sequencing data (163 mCRPC transcriptomes data from SU2C/PCF cohort), we found an inverse correlation between PSMA mRNA expression, and both BRCA2 mRNA expression (p < 0.001) and double-strand break repair (p < 0.001). This would suggest that high PSMA expression in mCRPC is associated with BRCA2 loss and a reduction in homologous recombination dependent DNA repair.
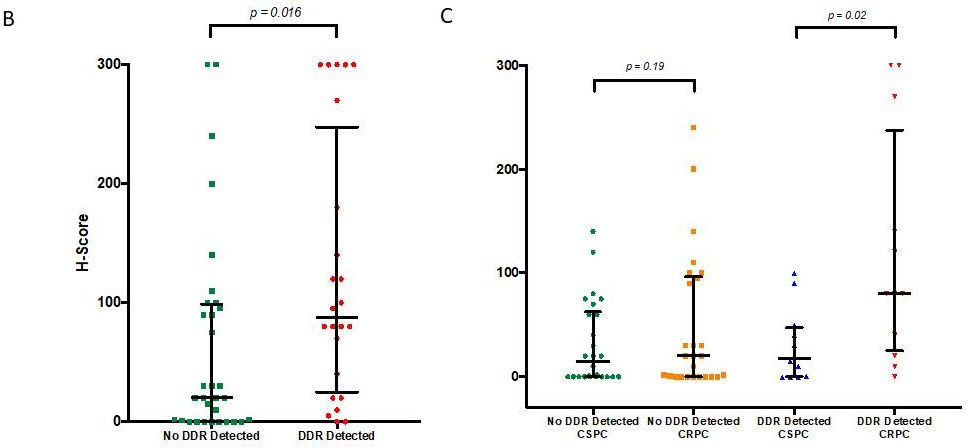
Figure 1B: Levels of mPSMA expression in patients with deleterious aberrations in genes involved in the DNA repair vs. those without
Figure 1C: Expression of mPSMA in matched mCRPC and CSPC in patients with detected deleterious DDR aberrations vs. patients without DDR aberrations
Conclusions
Taken together, these data indicate that a proportion of patients exhibit no detectable PSMA expression in their tumour biopsies at diagnosis, and this correlates with survival. Moreover, while mPSMA increases in mCRPC, a proportion of patients exhibit no detectable PSMA. When it is present, mPSMA expression exhibits intra and interpatient heterogeneity, with a potential impact on the clinical utility of PSMA theranostics.
Finally, we showed that deleterious DDR aberrations are associated with significantly higher mPSMA expression levels in mCRPC. Therefore, DDR biomarkers can potentially be used to guide PSMA-directed therapies.
The reference list can be made available to interested readers upon request by sending an email to: communications@uroweb.org.

